Results¶
The results section of the PMS tutorial does a good job walking you through the project directory structure for any HTPolyNet run, so we’ll skip that here.
First, if we look in proj-0/plots, we can see the density vs. time of the densification stage:
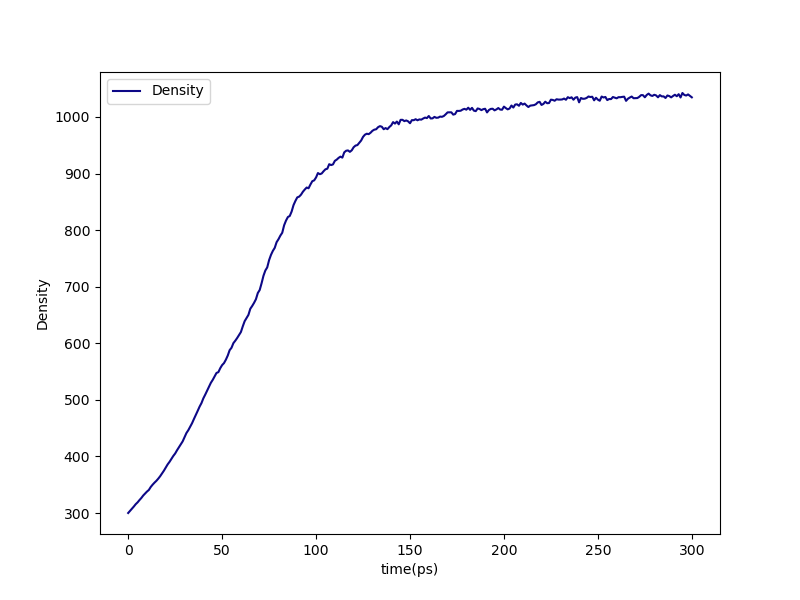
Fig. 15 Density vs. time during the densification stage of the DGEBA/PACM liquid.¶
As in the polymethylstyrene tutorial, we can make plots of the conversion vs. run time and the cure iteration vs. run time:
$ htpolynet plots diag --diags diagnostics.log
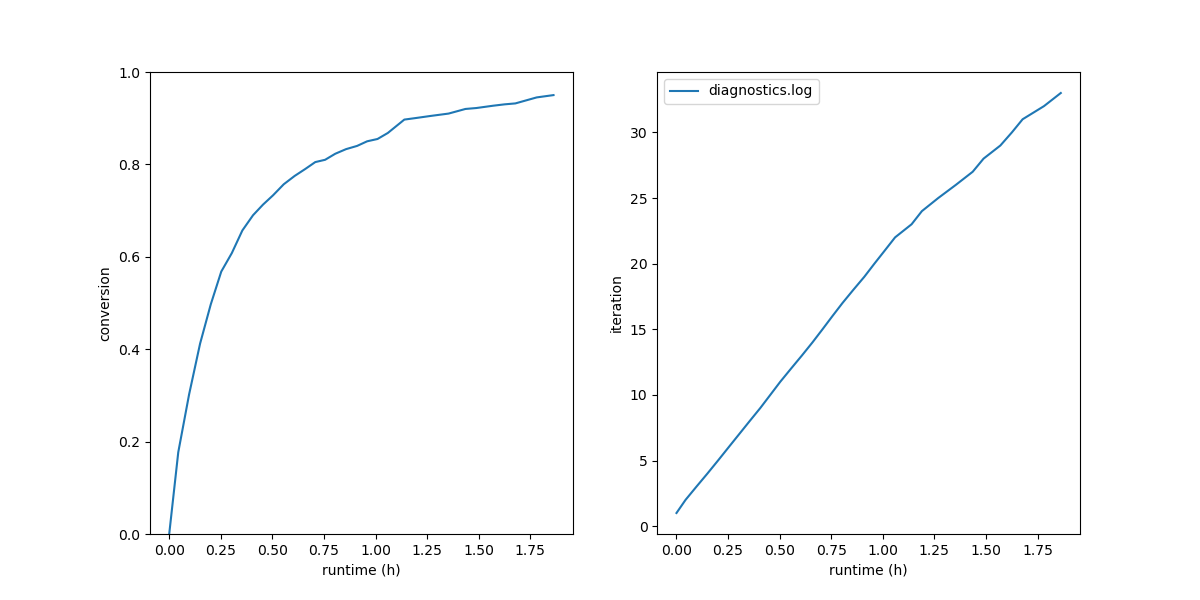
Fig. 16 (Left) Conversion vs. wall-clock time; (right) Iteration number vs wall-clock time.¶
This build was run on a standalone workstation with a very outdated GPU, so it is not surprising it took almost two hours. The behavior is typical of a build like this: about 80% cure is achieved in the first hour of run time, and then another hour or so is needed to get that last 20%.
Let’s generate traces of temperature, density, and potential energy vs time for the entire course of the system build:
$ htpolynet plots build --proj proj-0 --buildplot t --traces t d p
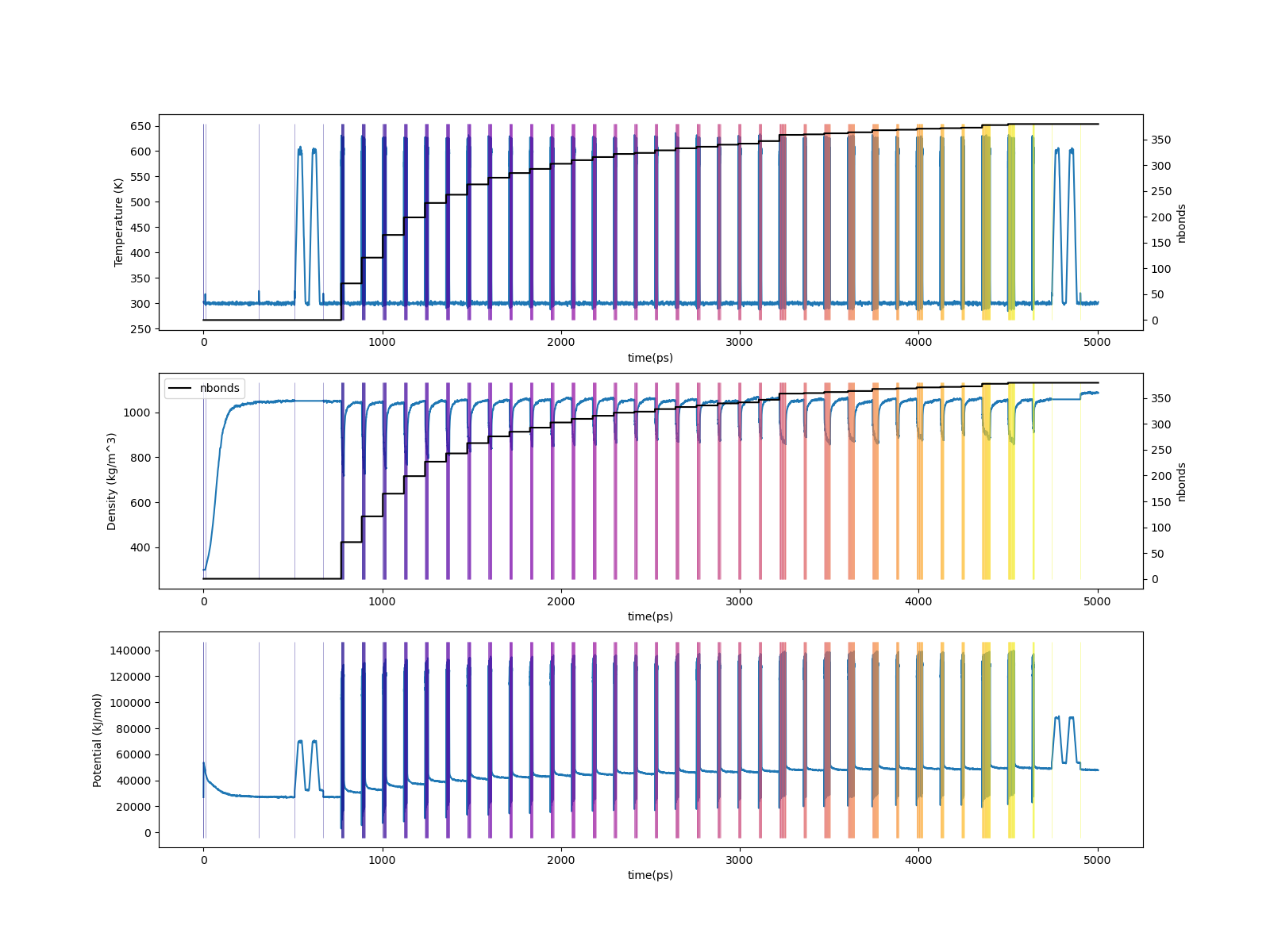
Fig. 17 (Top) Temperature vs. time for 95% cure of DGEBA/PACM; (Middle) Density vs time. The two topmost plots also report number of bonds; (Bottom) Potential energy vs. time.¶
Below we show before and after pictures of the system, where all bonds associated with crosslink sites are rendered in “licorice” with the rest in lines, and DGEBA molecules are mauve while PACMs are green:
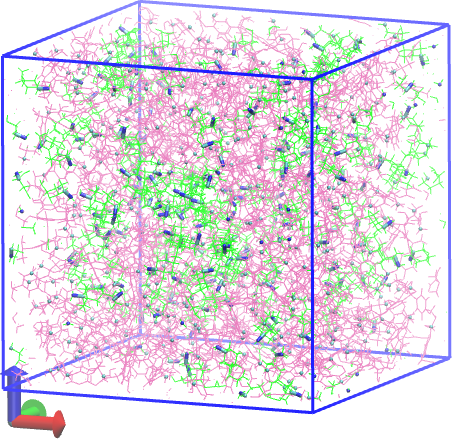
Fig. 18 System before cure.¶ |
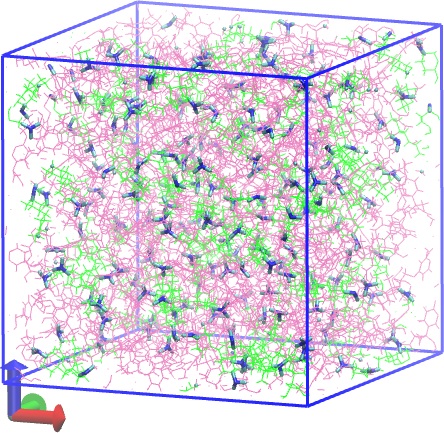
Fig. 19 System after cure.¶ |