Usage¶
Installation of the HTPolyNet package gives access to the htpolynet command. Invoking htpolynet --help shows basic usage of the command. Normal usage requires issuing a subcommand.
$ htpolynet <subcommand> <subcommand-arguments>
The following subcommands are available:
Subcommand |
Description |
|---|---|
|
Perform a polymerization or “build a project” |
|
Perform only a GAFF-parameterization of monomers and oligomers |
|
Report information about the |
|
Fetches examples provided in the package |
|
Perform some optional basic checks before launching a build |
|
Perform one or more post-build MD simulations |
|
Generate plots from output of builds or post-build simulations |
|
Peform |
htpolynet <subcommand> -h provides subcommand-level help.
Typical Usage¶
HTPolyNet is designed to allow one to easily simulate many system replicas of a given polymerization recipe. A typical use of HTPolyNet to measure thermomechanical properties of a polymerized system is shown below.
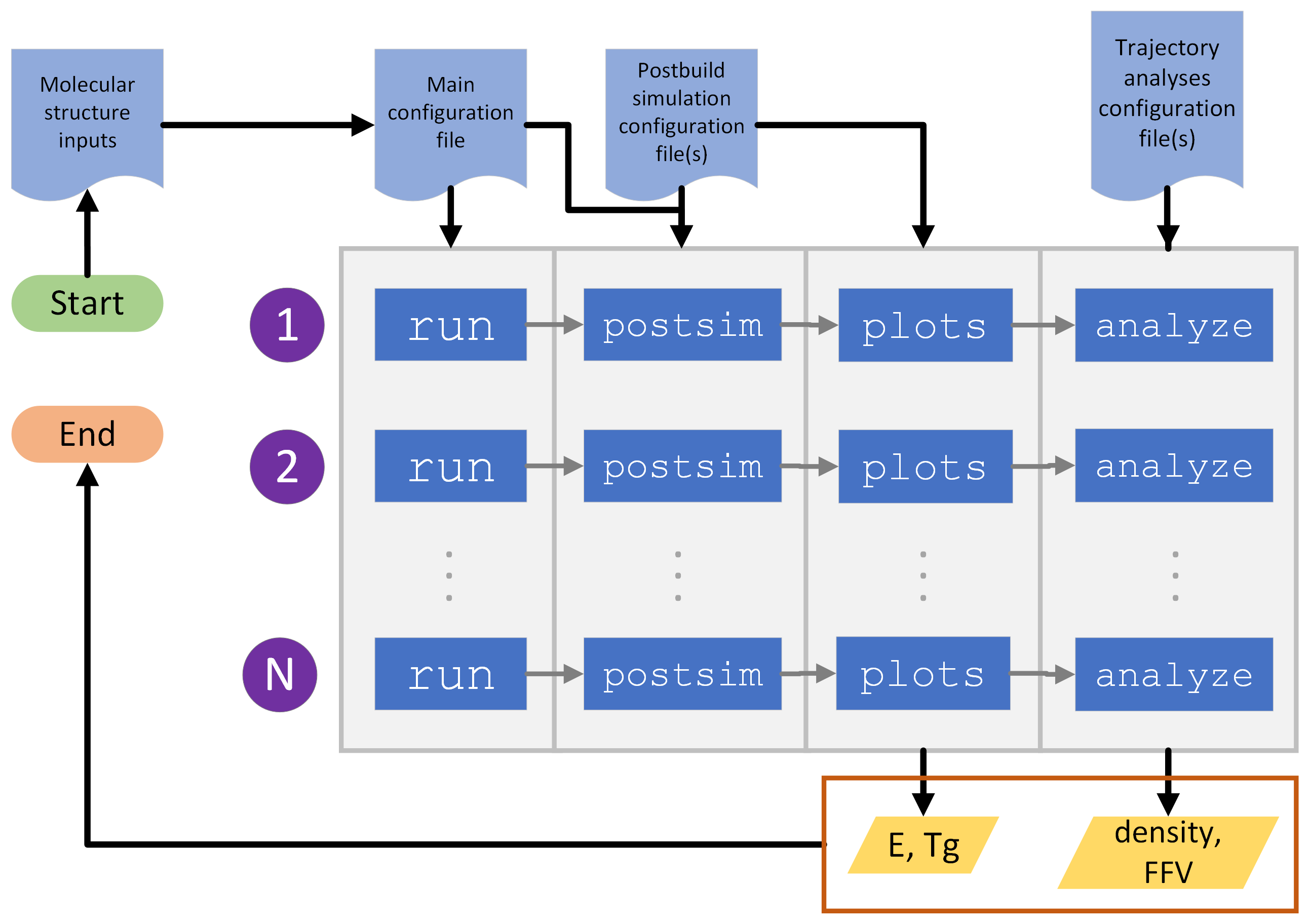
Fig. 1 Example of a typical workflow for using HTPolyNet to measure a system density, fractional free volume, Young’s modulus E and glass-transition temperature Tg.¶
One typically begins in a clean base directory, and generates any necessary input molecular structure files, either in mol2 or pdb format. One then creates the main configuration file for the polymerization. One can then launch as many instances of htpolynet run as one likes; each creates a unique system. Once all the systems are built, one can then use htpolynet postsim to run post-build MD simulations, and then use htpolynet plots to extract E and Tg. If one of the post-build simulations is a long equilibration, one can then use htpolynet analyze to extract observables from that simulation’s trajectory file; here, we indicate that one can extract density and fractional free volume.
The HTPolyNet Directory Structure¶
The htpolynet command is typically issued inside of a base directory. I organize my simulations so that each particular system composition and cure protocol I am considering goes in its own base, and multiple runs in that base are all just replica simulations for conducting sample averages. Each such simulation is referred to as a project and each has its own project directory generated by htpolynet run.
A base directory can just be an empty directory to start with. I usually initialize a base directory by giving it a molecule library:
.
└── lib
└── molecules
├── inputs
│ └── STY.mol2
└── parameterized
STY.mol2 is a mol2 file describing styrene in this example. HTPolyNet will store its parameterization information for all molecules and oligomers in the parameterized subdirectory.
htpolynet run will generate one or more project directories in the base directory.
HTPolyNet Subcommand Summaries¶
htpolynet run¶
run is used to generate a polymerized system (“build a project”) given directives in the configuration file and any required input monomer structures.
$ htpolynet run <config> [-lib <lib>] [-proj <projname>] \
[-diag <diagfile>] [--loglevel LOGLEVEL] [--restart] \
[--force-parameterization] [--force-checkin]
<config>refers to the name of the configuration input file.-libnames a directory that is treated as a local library of molecular structures. By default, this is assumed to be./lib/(that is, the directory you are in when you issuehtpolynet runis expected by default to have alib/directory; if not, theLibrarysubpackage of theHTPolyNetpackage will be queried for any data). At the beginning of a new run,lib/should have one subdirectory calledmolecules. Undermoleculesshould be the two directoriesinputsandparameterized.HTPolyNetwill look for inputmol2orpdbfiles inlib/molecules/inputs, and “check-in” the results of parameterized molecules (i.e., Gromacs formatgro,itp, andtopfiles) inlib/molecules/parameterized.-projnames the project directory. If not specified, or provided with the valuenext(the default),HTPolyNetwill create the next auto-named project directory. These are always named asproj-0,proj-1,proj-2, etc. If no project directory exists and an explicit one is not specified by-proj,HTPolyNetcreates the first one,proj-0:. ├── lib │ └── molecules │ ├── inputs │ │ └── STY.mol2 │ └── parameterized │ └── proj-0 ├── molecules │ └── parameterized ├── plots └── systems
The subdirectories of a project directory upon its creation are as follows.
molecules/parameterized– all molecular parameterization results appear here (in addition to being checked in to the library)
systems– system initializations, equilibrations, CURE iterations, and postcure equilibrations all get their own subdirectories here.
plots– various plots generated on the fly.
These will be explained more fully in the tutorials.
-diagnames the diagnostic output file, and--loglevelspecifies the logging level it uses. The default level isdebug(i.e., the most informative).-restartindicates that this is a restart (experimental!).--force-parameterizationsignals thatHTPolyNetshould perform all molecular parameterizations from scratch even if parameterizations exist in the library.--force-checkinsignals that any parameterizationsHTPolyNetperforms should have their results “checked-in” to the library, even if previous parameterizations are there already.
htpolynet parameterize¶
parameterize is the command for only performing molecular parameterizations and checking the results into a library:
$ htpolynet parameterize [-h] [-lib LIB] [-diag DIAG] [-restart] \
[--force-parameterization] [--force-checkin] [--loglevel LOGLEVEL] config
The command-line options of htpolynet parameterize have all the same meanings as they do for htpolynet run. The only difference is that htpolynet parameterize only performs the parameterization of all monomers and oligomer templates. The intention is that later invocations of htpolynet run can use these parameterizations without having to reperform them. Of course, since a first invocation of htpolynet run also peforms parameterizations and saves their results, it is strictly never necessary to use htpolynet parameterize. However, if your parameterizations have issues, it is cleaner to use htpolynet parameterize to try to fix them.
htpolynet info¶
This simply outputs some information about HTPolyNet.
$ htpolynet info
This is some information on your installed version of HTPolyNet
System library is /home/cfa/Git/HTPolyNet/Library
Ambertools:
antechamber (ver. 22.0) at antechamber
tleap (ver. 22.0) at tleap
parmchk2 (ver. 22.0) at parmchk2
htpolynet info only reports the absolute pathname of the Library subpackage for your reference, and the fully resolved command names for the three required Ambertools executables antechamber, tleap and parmchk2, along with their versions. If they are already in your path, the results appear as above.
htpolynet plots¶
If invoked inside of a base directory containing one or more project directories, plots instructs HTPolyNet to generate some plots.
$ htpolynet plots --help
usage: htpolynet plots [-h] [--diags DIAGS [DIAGS ...]] [--proj PROJ [PROJ ...]] \
[--cfg CFG [CFG ...]] [--buildplot {t,g,n,c} [{t,g,n,c} ...]] \
[--traces {t,d,p} [{t,d,p} ...]] [--n_points N_POINTS N_POINTS] \
[--plotfile PLOTFILE] [--no-banner] [--loglevel LOGLEVEL] \
{diag,build,post}
positional arguments:
{diag,build,post} source of data to plot
"diag" takes data from any diagnostic output
"build" takes data the console output
"post" uses a ``postsim`` configuration file
options:
-h, --help show this help message and exit
--diags DIAGS [DIAGS ...]
names of diagnostic log files (1 or more)
--proj PROJ [PROJ ...]
name of project director[y/ies]
--cfg CFG [CFG ...] name input config files
--buildplot {t,g,n,c} [{t,g,n,c} ...]
type of build plot to generate: t: traces (select using --traces); g: 2-D graph
representations iteration by iteration; n: homo-N between crosslinks; c: cluster-size
distributions
--traces {t,d,p} [{t,d,p} ...]
type of traces to plot from build: t: temperature; d: density; p: potential energy
--n_points N_POINTS N_POINTS
number of [cold-side,hot-side] data points in the Tg analysis to fit lines to
--plotfile PLOTFILE name of plot file to generate
--no-banner turn off the banner
--loglevel LOGLEVEL Log level for messages written to diagnostic log (debug|info)
The plots subcommand manages plot generation from data from any of three sources: diagnostics, build, or post-build MD simulations. (Post-build MD simulations are described below for the htpolynet postsim subcommand.) We demonstrate examples of all three types in the tutorials.
htpolynet fetch-example¶
This will fetch one or more examples from the example_depot of the system Library.
$ htpolynet fetch-example --help
usage: htpolynet fetch-example [-h] [-n {0,1,2,3,4,5,6,all}] [-k]
options:
-h, --help show this help message and exit
-n {0,1,2,3,4,5,6,all}
number of example tarball to unpack from 0-liquid-styrene, 1-polystyrene,
2-polymethylstyrene, 3-bisgma-styrene-thermoset, 4-pacm-dgeba-epoxy-thermoset,
5-dfda-fde-epoxy-thermoset,
6-htpb-ipdi
-k keep tarballs
Fetching will copy the tarball for the requested system to the current directory and then untar it and remove it, leaving behind the directory. For example to fetch the PACM-DGEBA epoxy thermoset example:
$ htpolynet fetch-example -n 4
$ ls
4-pacm-dgeba-epoxy-thermoset/
$ cd 4-pacm-dgeba-epoxy-thermoset
$ ls
DGE-PAC-hi.yaml DGE-PAC-lo.yaml lib/ run.sh
This folder (like all example folders) comes with two configuration files that differ only the the requested degree of cure. “hi” refers to 95% cure, and “lo” to 50%. Also provided is the ./lib/molecules folders with the ./lib/molecules/inputs and ./lib/molecules/parameterized empty subfolders. Finally, the bash script run.sh can just be invoked to build the input monomers and run the two builds in series. This will be described in much more detail in the tutorials.
htpolynet fetch-example -n all just grabs all seven examples.
htpolynet input-check¶
The purpose of this subcommand is to report the size of the initial system that would be created by the provided configuration file and monomer input structures.
$ htpolynet input-check DGE-PAC-hi.yaml
Molecule DGE: 53 atoms, 200 molecules
Molecule PAC: 41 atoms, 100 molecules
DGE-PAC-hi.yaml: 14700 atoms in initial system
htpolynet postsim¶
The purpose of the postsim subcommand is to control the execution of several types of post-build MD simulations.
$ htpolynet postsim -h
usage: htpolynet postsim [-h] [-proj PROJ [PROJ ...]] [-lib LIB] \
[-ocfg OCFG] [-cfg CFG] [--no-banner] [--loglevel LOGLEVEL]
options:
-h, --help show this help message and exit
-proj PROJ [PROJ ...]
name of project directory
-lib LIB local user library of molecular structures and parameterizations
-ocfg OCFG original HTPolyNet config file used to generate project(s)
-cfg CFG config file for specifying the MD simulations to perform
--no-banner turn off the banner
--loglevel LOGLEVEL Log level for messages written to diagnostic log (debug|info)
The simulations are controlled by the YAML-format config file, which is _distinct_ from the cfg file used to generate the project(s). The types of post-build MD simulations available are annealing, equilibration, temperature-ladder, and uniaxial deformation. Demonstrations of how the ladder and deformation simulations are used to compute the glass-transition temperature and Young’s modulus are detailed in the tutorials.
htpolynet analyze¶
The analyze subcommand provides a shortcut interfaces to selected gmx-style trajectory analysis calculations.
usage: htpolynet analyze [-h] [-proj PROJ [PROJ ...]] [-lib LIB] [-ocfg OCFG] [-cfg CFG] [--no-banner] [--loglevel LOGLEVEL]
options:
-h, --help show this help message and exit
-proj PROJ [PROJ ...]
name of project directory
-lib LIB local user library of molecular structures and parameterizations
-ocfg OCFG original HTPolyNet config file used to generate project(s)
-cfg CFG config file for specifying the analyses to perform
--no-banner turn off the banner
--loglevel LOGLEVEL Log level for messages written to diagnostic log (debug|info)
The arguments of analyze are similar to those of postsim, except the configuration file specifies the analysis to perform, and the -proj specifies which project directories to perform the analysis in. We demonstrate using htpolynet analyze in the tutorials.