Reactions¶
There are three types of reactions defined in the YAML configuration file. The first two describe how to build a bisGMA molecule, the next four describe the polymerization reactions, and the final two describe capping reactions for unreacted monomers.
Building bisGMA¶
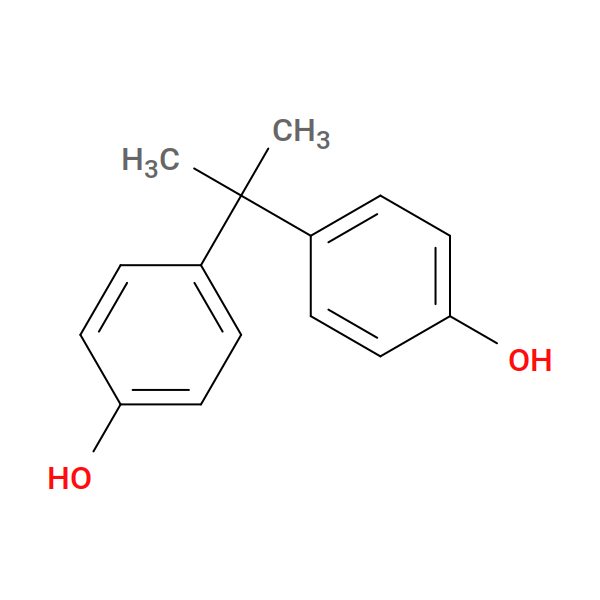
Fig. 20 Bisphenol-A¶ |
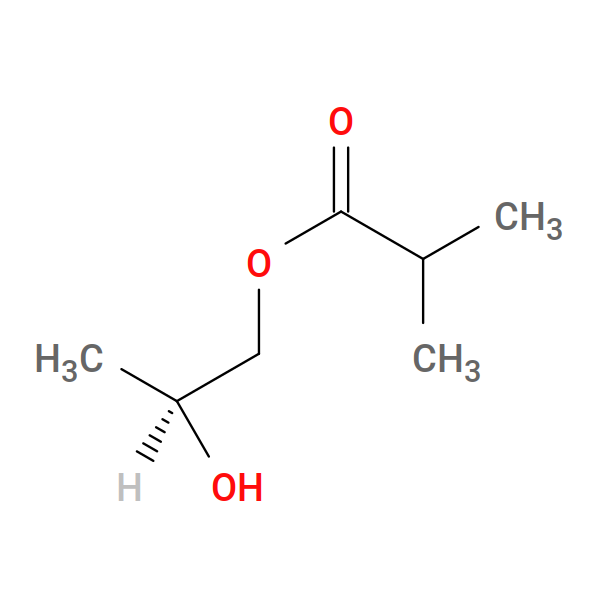
Fig. 21 2-hydroxypropyl isopropyl ester (HIE)¶ |
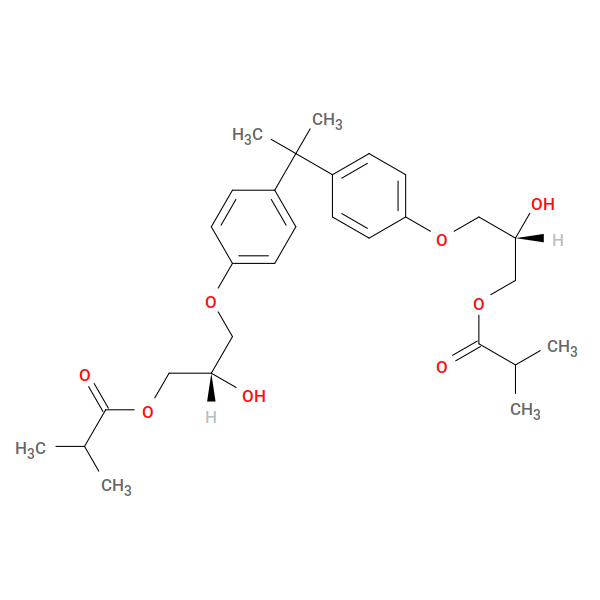
Fig. 22 bis GMA (active form)¶ |
Let’s focus first on the reactions used to build bis GMA. First, we react one HIE to one hydroxyl on BPA:
- name: B1
stage: param
reactants:
1: BPA
2: HIE
product: GM1
atoms:
A:
reactant: 1
resid: 1
atom: O1
z: 1
B:
reactant: 2
resid: 1
atom: C4
z: 2
bonds:
- atoms: [A, B]
order: 1
This creates the intermediate GM1, which we then use to in the second reaction:
- name: B2
stage: param
reactants:
1: GM1
2: HIE
product: GMA
atoms:
A:
reactant: 1
resid: 1
atom: O2
z: 1
B:
reactant: 2
resid: 1
atom: C4
z: 1
bonds:
- atoms: [A, B]
order: 1
Note that both of these reactions are stage “param”, since we know we need to GAFF-parameterize the results to arrive at a fully parameterized GMA molecule.
Why are we doing this instead of just making a full GMA from scratch using SMILES and obabel, or drawing the darn thing in ChemSketcher? The reason is that the HIE monomers house the reactive C=C double bonds, and the two HIE monomers in one GMA are to be treated as equivalent. This means that we need to specify cure reactions between STY and HIE rather than between STY and GMA. This will greatly reduce the size of the expanded reaction set.
Cure reactions¶
Now that the input structures for BisGMA and styrene have been generated, we are ready to generate the reaction dictionaries that describe the chemistry. BisGMA has two carbon-carbon double bonds, each housed in HIE monomers between their C1 and C2 atoms. As on styrene, we have also chosen on HIE to let C1 be the radical carbon and C2 be the methyl.
Our basic reaction model is that a radical carbon “attacks” a methyl carbon, ejecting 2 sacrifical H’s and forming a single C-C bond. We must therefore encode four basic reaction scenarios:
Attacker (C1 owner) |
Attackee (C2 owner) |
|---|---|
HIE |
HIE |
styrene |
styrene |
HIE |
styrene |
styrene |
HIE |
For each scenario above, we need only encode the C1-attacks-C2 reaction.
- name: dimer_xx
stage: cure
reactants:
1: STY
2: STY
product: STY~C1-C2~STY
atoms:
A:
reactant: 1
resid: 1
atom: C1
z: 1
B:
reactant: 2
resid: 1
atom: C2
z: 1
bonds:
- atoms: [A, B]
order: 1
- name: dimer_yy
stage: cure
reactants:
1: HIE
2: HIE
product: HIE~C1-C2~HIE
atoms:
A:
reactant: 1
resid: 1
atom: C1
z: 1
B:
reactant: 2
resid: 1
atom: C2
z: 1
bonds:
- atoms: [A, B]
order: 1
- name: dimer_xy
stage: cure
reactants:
1: STY
2: HIE
product: STY~C1-C2~HIE
atoms:
A:
reactant: 1
resid: 1
atom: C1
z: 1
B:
reactant: 2
resid: 1
atom: C2
z: 1
bonds:
- atoms: [A, B]
order: 1
- name: dimer_yx
stage: cure
reactants:
1: HIE
2: STY
product: HIE~C1-C2~STY
atoms:
A:
reactant: 1
resid: 1
atom: C1
z: 1
B:
reactant: 2
resid: 1
atom: C2
z: 1
bonds:
- atoms: [A, B]
order: 1
Finally, we can include capping reactions that revert any completely unreacted double bonds back to actual double-bonds (notice the order specifications):
- name: styCC
stage: cap
reactants:
1: STY
product: STYCC
probability: 1.0
atoms:
A:
reactant: 1
resid: 1
atom: C1
z: 1
B:
reactant: 1
resid: 1
atom: C2
z: 1
bonds:
- atoms: [A, B]
order: 2
- name: hieCC
stage: cap
reactants:
1: HIE
product: HIECC
probability: 1.0
atoms:
A:
reactant: 1
resid: 1
atom: C1
z: 1
B:
reactant: 1
resid: 1
atom: C2
z: 1
bonds:
- atoms: [A, B]
order: 2
As we will detail in the next section, there are no symmetry-equivalent atoms necessary in this system, even though bisGMA is a symmetric molecule. This is because we need only address HIE monomers in the cure reactions; BPA’s are “silent” with respect to cure. There are no symmetry-equivalent atoms within a single HIE monomer.
However, because this is a system of double-bonds that open to react in each monomer, we do have to allow HTPolyNet to enumerate all possible combinations of -(C1-C2)- chains of length 3 and 4 to fully cover all template needs. This is referred to as chain expansion of reactions. For all length-3 chains, there are 8 possible sequences of STY and HIE, and for each, the actual bond we focus on can be in one of two positions in that chain (between the first and second, or between the second and third), so there are 16 distinct trimers in the chain-expanded reaction set. Likewise, there are 16 possible chains of length 4, but we need only condsider those for which the bond that forms is between the second and third, since trimer parameterizations cover the other two positions. This gives a total of 32 cure reactions in the chain-expanded set of reactions.
The 16 trimers are:
STY~C1=C2~STY~C1-C2~STY
HIE~C1=C2~STY~C1-C2~STY
HIE~C1=C2~HIE~C1-C2~HIE
HIE~C1=C2~STY~C1-C2~HIE
HIE~C1=C2~HIE~C1-C2~STY
STY~C1=C2~STY~C1-C2~STY
STY~C1=C2~HIE~C1-C2~HIE
STY~C1=C2~STY~C1-C2~HIE
STY~C1=C2~HIE~C1-C2~STY
STY~C1-C2~STY~C1=C2~HIE
HIE~C1-C2~HIE~C1=C2~HIE
STY~C1-C2~HIE~C1=C2~HIE
HIE~C1-C2~STY~C1=C2~HIE
STY~C1-C2~STY~C1=C2~STY
HIE~C1-C2~HIE~C1=C2~STY
STY~C1-C2~HIE~C1=C2~STY
HIE~C1-C2~STY~C1=C2~STY
The = signifies the single C-C bond that is the focus of the parameterization template; note that the sequences of the first 8 are repeated in the second 8; only the position of the templated bond is different between the two sets.
The 16 quadrimers are:
STY~C1-C2~STY~C1=C2~STY~C1-C2~STY
STY~C1-C2~STY~C1=C2~HIE~C1-C2~HIE
STY~C1-C2~STY~C1=C2~STY~C1-C2~HIE
STY~C1-C2~STY~C1=C2~HIE~C1-C2~STY
HIE~C1-C2~HIE~C1=C2~STY~C1-C2~STY
HIE~C1-C2~HIE~C1=C2~HIE~C1-C2~HIE
HIE~C1-C2~HIE~C1=C2~STY~C1-C2~HIE
HIE~C1-C2~HIE~C1=C2~HIE~C1-C2~STY
STY~C1-C2~HIE~C1=C2~STY~C1-C2~STY
STY~C1-C2~HIE~C1=C2~HIE~C1-C2~HIE
STY~C1-C2~HIE~C1=C2~STY~C1-C2~HIE
STY~C1-C2~HIE~C1=C2~HIE~C1-C2~STY
HIE~C1-C2~STY~C1=C2~STY~C1-C2~STY
HIE~C1-C2~STY~C1=C2~HIE~C1-C2~HIE
HIE~C1-C2~STY~C1=C2~STY~C1-C2~HIE
HIE~C1-C2~STY~C1=C2~HIE~C1-C2~STY
So, to review: we needed only to specify the four possible polymerization reactions involving STY and HIE monomers, and HTPolyNet automatically takes care of generating all relevant product templates that could be needed during any curing system.
The next thing we consider is the configuration file necessary to describe the crosslinking chemistry and determine the system build.